| Drug Design Laboratory |
|
 |
| Sitemap |
| Print Version |
| Login |
| About CMSimple |
| News > Software projects > AMMP VE |
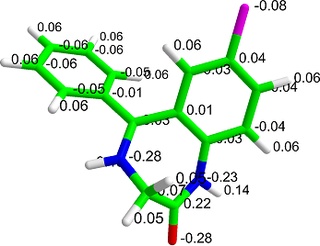
AMMP VEAnother Molecular Mechanics Program
AMMP is a modern full-featured molecular mechanics, dynamics and modelling program. It can manipulate both small molecules and macromolecules including proteins, nucleic acids and other polymers. In addition to standard features, like numerically stable molecular dynamics, fast multipole method for including all atoms in the calculation of long range potentials and robust structural optimizers, it has a flexible choice of potentials and a simple yet powerful ability to manipulate molecules and analyze individual energy terms. One major advantage over many other programs is that it is easy to introduce non-standard polymer linkages, unusual ligands or non-standard residues. Adding missing hydrogen atoms and completing partial structures, which are difficult for many programs, are straightforward in AMMP. It is written in C and has been ported to many different computers. The source code is available under the GNU "copyleft" so that it is easy to follow exactly what the program is doing. Another major advantage over commercial software is that AMMP can be easily included in another program to supply molecular mechanics function.
AMMP VE (VEGA Edition) is a modified version of the original AMMP project developed by Robert W. Harrison (for more information, see the original Web site). The new features included in the VE edition
Download Current release: 2.4.0 The Windows 32 and 64 bit versions are included in the latest VEGA ZZ package and so if you want to use these operating systems, it's strongly recommended to download that distribution (for more details, go to the VEGA ZZ main site). Included in this package:
|
| < | Privacy and cookies | > |