6.16 Build a molecule with SMILES
VEGA ZZ can build molecules from their
SMILES strings, selecting Edit
![]() Build
Build
![]() SMILES in the main menu.
The 1D structure of the SMILES string is converted to 3D clicking the Build
button. You can select Current workspace or New workspace
as Destination of the 3D structure.
SMILES in the main menu.
The 1D structure of the SMILES string is converted to 3D clicking the Build
button. You can select Current workspace or New workspace
as Destination of the 3D structure.
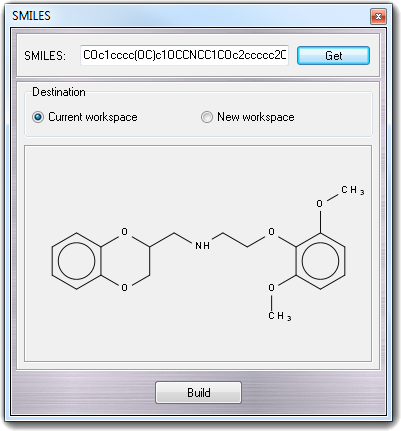
When you edit the SMILES string, the 2D preview is automatically updated showing the changes.
If the current workspace is not empty, the Get button allows you to convert
the 3D structure to a SMILES string. The same operation is possible by selecting
Edit
![]() Copy special in the main
menu and by choosing SMILES, but the resulting string is copied to the
clipboard instead of the SMILES window.
Copy special in the main
menu and by choosing SMILES, but the resulting string is copied to the
clipboard instead of the SMILES window.
6.16.2 Combinatorial SMILES builder
If you need to build a large number of molecules sharing the same scaffold, you
can use the combinatorial SMILES builder, whose dialog window can be opened by
selecting Edit
![]() Build
Build
![]() Combi SMILES in the main menu. As in the standard SMILES builder, you can
put directly the scaffold structure in the SMILES scaffold field or you
can obtain it from the current workspace by clicking Get button. For
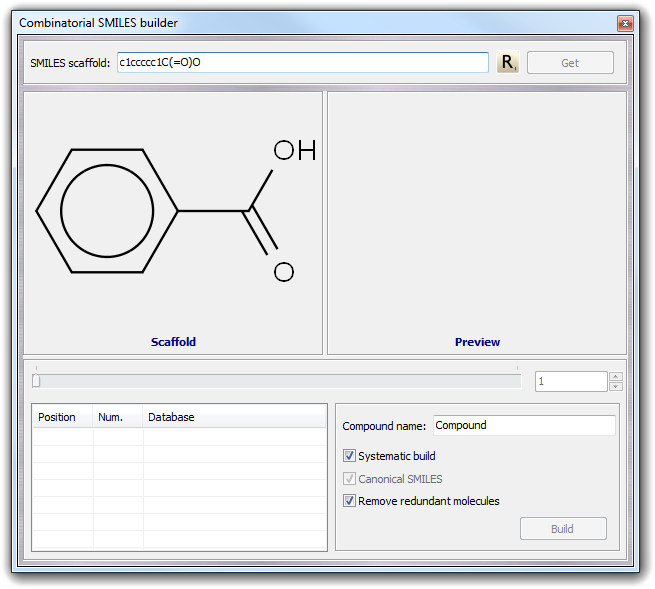
example, imagine to build a set of derivatives of the benzoic acid:
Combi SMILES in the main menu. As in the standard SMILES builder, you can
put directly the scaffold structure in the SMILES scaffold field or you
can obtain it from the current workspace by clicking Get button. For
example, imagine to build a set of derivatives of the benzoic acid:
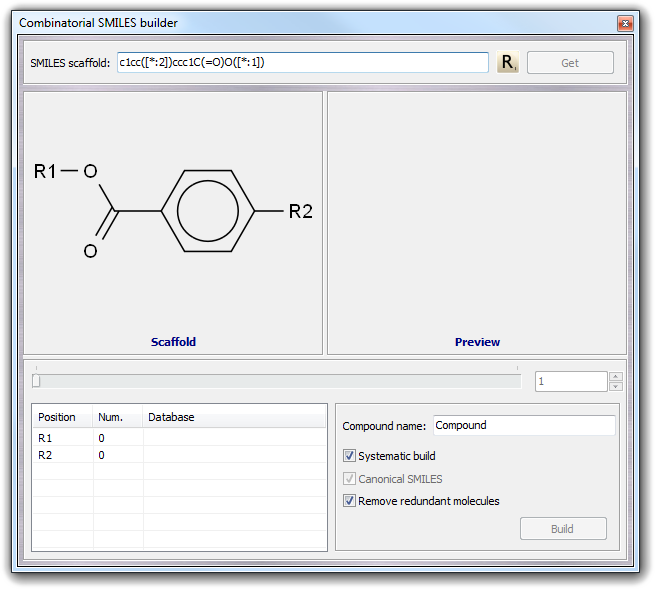
To insert the variable positions in which the residues will be added, you must move the cursor in the SMILES string and click R button as shown below:
The next step is to add one or more SMILES databases to each variable position (R1 and R2 in this case). You can use a custom fragment database built by you (for more information about the SMILES database format, click here) by double clicking on each position row, by the context menu (Open item) and by drag & drop of the files over the position in the table. Alternatively, you can add the built-in databases by using the context menu. To each position, you can add more than one database and if a position doesn't have any database, it is simply ignored.
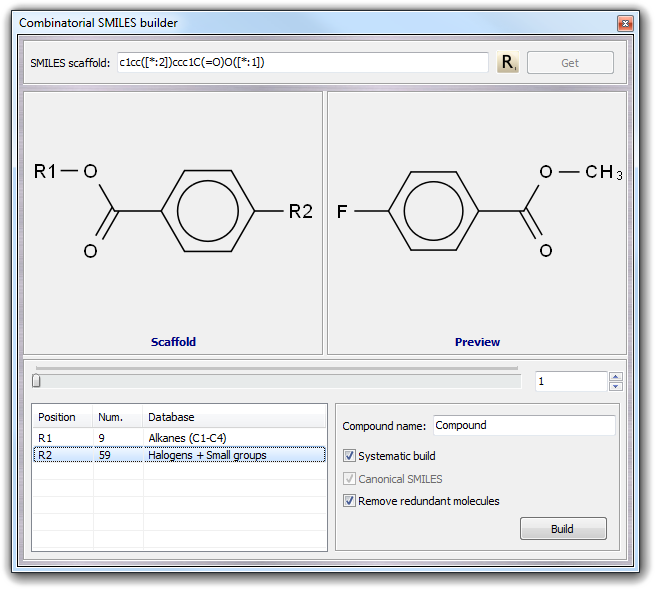
You can change the preview by moving the horizontal slider, by typing the compound number or by clicking the up and down buttons. The context menu of the table allows you to remove the fragments of a single position (Remove) or of all positions (Remove all). By the context menu of Scaffold and Preview boxes, you can copy the SMILES strings to the clipboard and print the structures.
In the bottom right box, you can change the name prefix that will be used for each compound (Compound name field) and some parameters as the type of build. If you check Systematic build, all possible fragment combination are considered, otherwise each fragment is considered only one time. In the case shown above, unchecking Systematic build, only 9 molecules are built that is the smallest number of fragment for a specific position. When you build a large number of derivatives, it may be possible to obtain non-unique molecules for symmetry reasons or due to duplicated fragments in different databases. To avoid this problem, you can check Remove redundant molecules.
The resulting compounds are saved in a SMILES database when you click Build button and if you want canonical SMILES strings, you can check Canonical SMILES.
Notes:
When you obtained the molecules by this tool, it may be needed to convert the resulting SMILES database to another format or to convert the molecules to 3D. You can do these operations in easy way by opening the SMILES database in the database explorer and copying the molecules to an empty database in the format that you need activating the 3D conversion in the Options tab.