12.3 ClustalX - Multiple Sequence Alignments
12.3.1 Introduction
This plug-in interfaces VEGA ZZ to ClustalX, NJPlot and Unrooted in order to performs multiple sequence alignments, profile analysis, philogenetic trees calculation and their visualization.
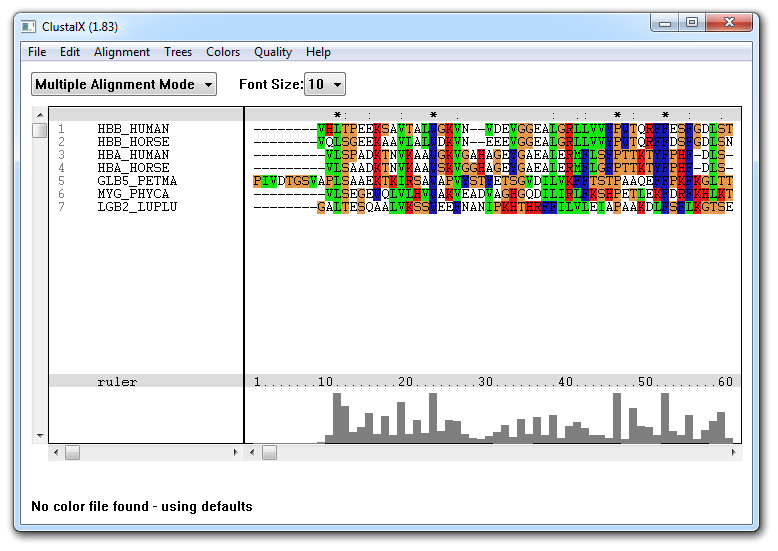
ClustalX provides a new window-based user interface to the
ClustalW multiple
alignment program. The sequence alignment is displayed in a window on the
screen. A versatile coloring scheme has been incorporated allowing you to
highlight conserved features in the alignment. The pull-down menus at the top of
the window allow you to select all the options required for traditional multiple
sequence and profile alignment. To open the main window, you must select
Bioinformatics
![]() Predator
from the main menu:
Predator
from the main menu:
For more information, please consult the ClustalX, ClustalV and ClustalW documentations in the ClustalX installation directory.
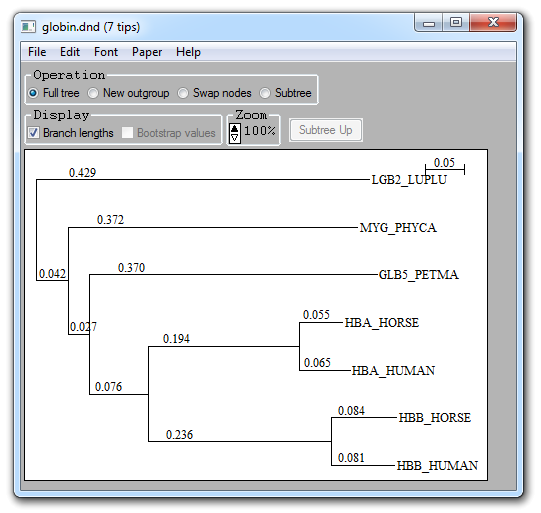
NJPlot and Unrooted are
tools for philogenetic tree visualization. The first displays rooted tree and
can be opened selecting Bioinformatics
![]() NJPlot:
NJPlot:
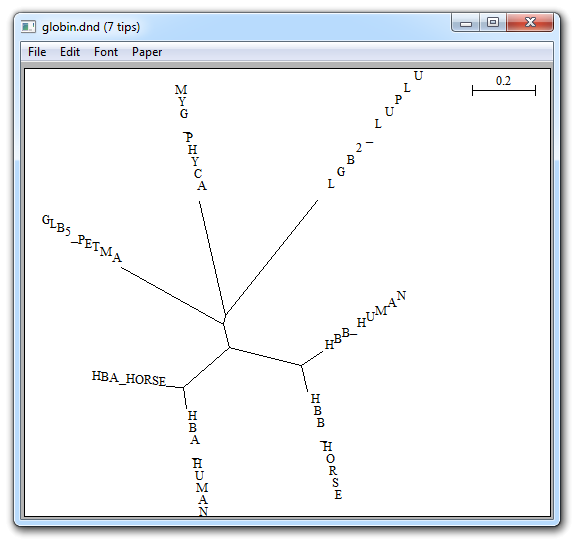
Unrooted, shown by Bioinformatics
![]() NJPlot unrooted, allows to display philogenetic trees in unrooted
mode:
NJPlot unrooted, allows to display philogenetic trees in unrooted
mode:
12.3.3.1 NJPlot original documentation
About NJPlot
Written by M. Gouy (mgouy@biomserv.univ-lyon1.fr).
Phylogenetic trees read from Newick formatted files can be displayed, re-rooted, saved, and plotted to PostScript (or PICT for the Macintosh) files.
Input trees can be with/without branch lengths, with/without bootstrap values, rooted or unrooted. Only binary trees are accepted.
NJPlot is available by anonymous FTP at pbil.univ-lyon1.fr in directory /pub/mol_phylogeny/njplot/ for MAC, PC under Windows95/NT and several unix
platforms.
njplot uses the Vibrant library by J. Kans.
Menu File
Open: To read a tree file in the Newick format (i.e., the file format used for trees by Clustal, PHYLIP and other programs).
Save plot: To save the tree plot in PostScript format, or in the PICT format for the Macintosh.
Save tree: To save the tree in a file with its current rooting.
Print: To print tree using the number of pages set by menu "Paper".
Menu Edit
Copy: [Mac & Windows ONLY!] Copies the current tree plot to the Clipboard
so that the plot can be pasted to another application.
Paste: If the clipboard contains a parenthesized tree, this tree will be plotted.
Clear: Clears the current plot so that new tree data can be pasted.
Find: To search for a taxon in tree and display it in red. Enter a (partial) name, case is not significant.
Again: Redisplay in red names matching the string entered in a previous Find operation.
Menu Font
Allows to change the font face and size used to display tree labels.
Menu Paper
Allows to set the paper size used by the "Save plot" and "Print" items of menu "File".
Pagecount (x): sets the number of pages used by "Save plot".
Operations
Full tree: Normal tree display of entire tree
New outgroup: Allows to re-root the tree. The tree becomes displayed with added # signs. Clicking on any # will set descending taxa as an outgroup
to remaining taxa.
Swap nodes: Allows to change the display order of taxa. The tree becomes displayed with added # signs. Clicking on any # will swap corresponding taxa.
Subtree: Allows to zoom on part of the tree. The tree becomes displayed with added # signs. Clicking on any # will limit display to descending taxa. Select "Show tree" to go back to full tree display.
Display
Branch lengths: If the tree contains branch lengths, they will be displayed. For readability, very short lengths are not displayed.
Bootstrap values: If the tree contains bootstrap values, they will be displayed.
Subtree up
When a subtree is being displayed, allows to add one more node towards the
root in the displayed tree part.
12.3.4 Copyright
ClustalX
Multiple Sequence Alignment Program
© Toby Gibson EMBL, Heidelberg, Germany
Des Higgins UCC, Cork, Ireland
Julie Thompson/Francois Jeanmougin IGBMC, Strasbourg, France.
NJPlot and Unrooted
Phylogenetic Trees Drawing Software
© Manolo Gouy, Lion, France