
![]() Run script in the main menu (for more information
click here), expand the script tree at
Docking, thus at PLANTS level and finally double click Patch bin
1.1.c.
Run script in the main menu (for more information
click here), expand the script tree at
Docking, thus at PLANTS level and finally double click Patch bin
1.1.c.

![]() Tool
bars item in Main menu.
Tool
bars item in Main menu.![]() Console menu item to show or hide it.
Console menu item to show or hide it.![]() 3D
controls.
3D
controls.
![]() Add
Add
![]() Atom it's possible to add one or more new
atoms:
Atom it's possible to add one or more new
atoms: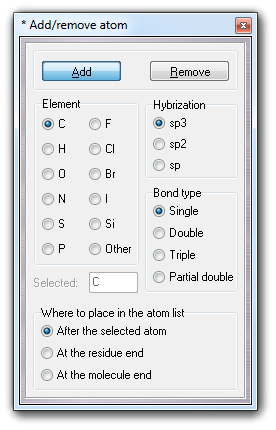
![]() Remove
Remove
![]() Atoms item in the main menu or
click the Remove button of Add/remove atom dialog box.
Atoms item in the main menu or
click the Remove button of Add/remove atom dialog box.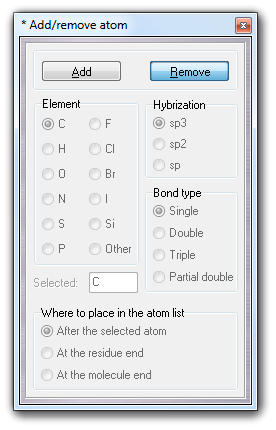
![]() Change
Change
![]() Atom/residue/chain
menu item.
Atom/residue/chain
menu item.![]() Change
Change
![]() Atom/residue/chain
and allows to change some atom, residue and chain
properties of the selected atom/molecule. For example, consider the following molecule
fragment:
Atom/residue/chain
and allows to change some atom, residue and chain
properties of the selected atom/molecule. For example, consider the following molecule
fragment: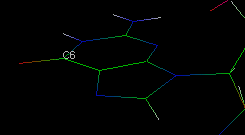
![]() Coordinates
Coordinates
![]() Constraints menu item:
Constraints menu item:


![]() Add
Add
![]() Centroid
in main menu:
Centroid
in main menu:
![]() Remove
Remove
![]() Centroids.
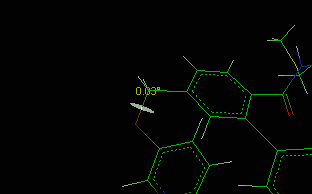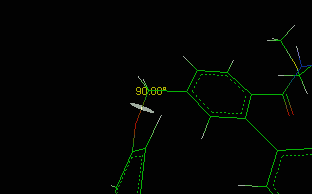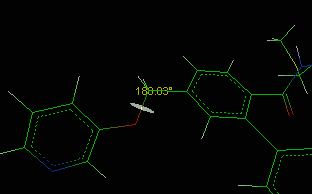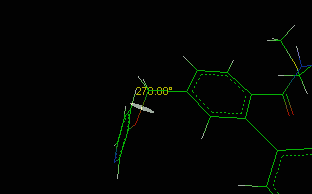
The centroids can be removed as normal atoms also. A typical application example
could be
the measure of the distance between two rings, as shown in the following
picture:
Centroids.
The centroids can be removed as normal atoms also. A typical application example
could be
the measure of the distance between two rings, as shown in the following
picture: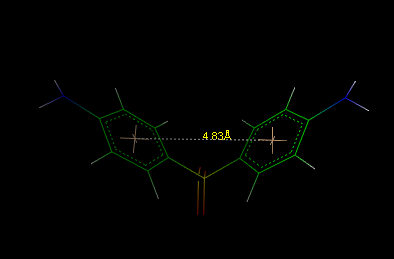
![]() Add
Add
![]() Bonds
in the main menu,
it's possible to add one or more new bonds:
Bonds
in the main menu,
it's possible to add one or more new bonds: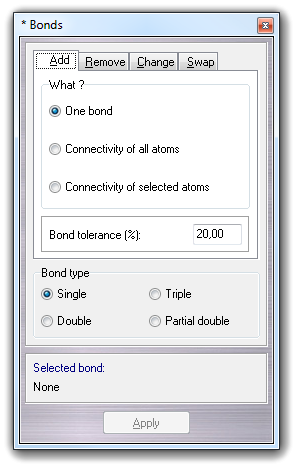
![]() Remove
Remove
![]() Bond
in the main menu.
Bond
in the main menu.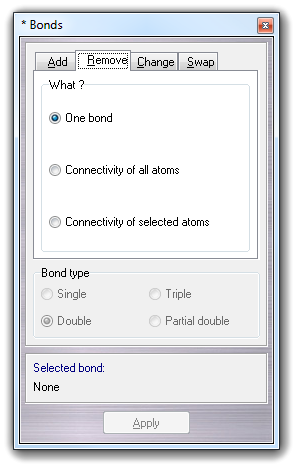
![]() Change
Change
![]() Bond
type
menu item:
Bond
type
menu item: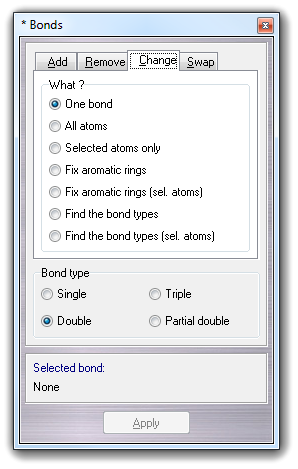
![]() Change
Change
![]() Swap bonds
menu item:
Swap bonds
menu item: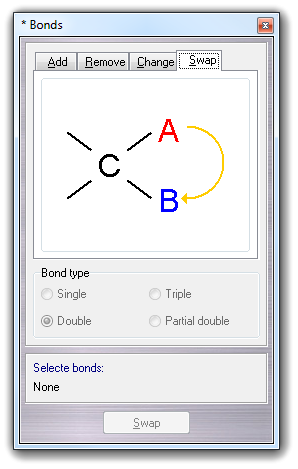
![]() Change
Change
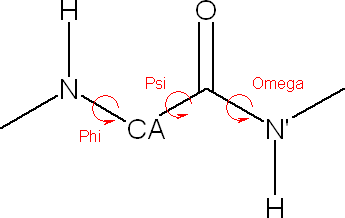
![]() Bond/Angle/Torsion item in the main menu.
Bond/Angle/Torsion item in the main menu.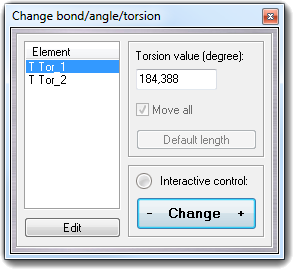
![]() Change
Change
![]() Secondary struct. in main menu.
Secondary struct. in main menu.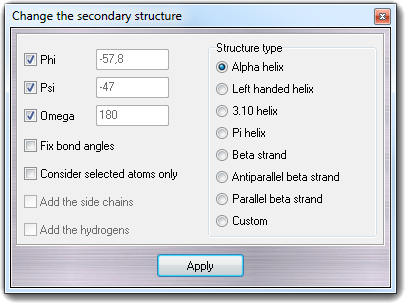
![]() Add
Add
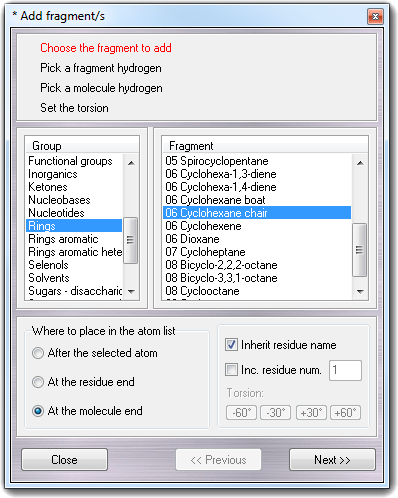
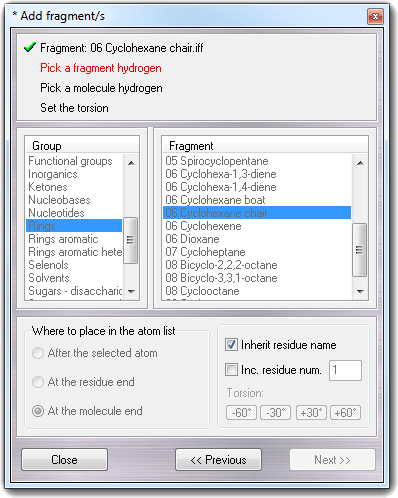
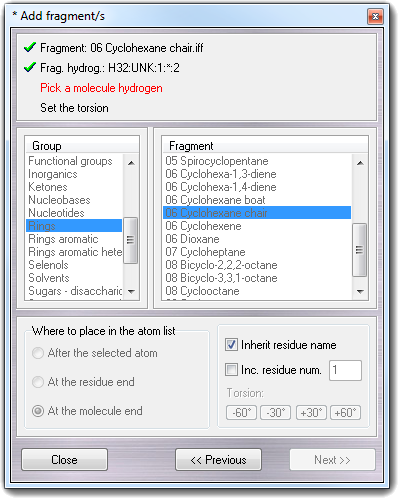
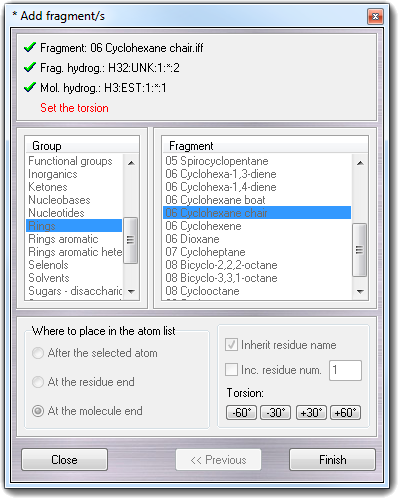
![]() Fragments menu item, the following
wizard is shown:
Fragments menu item, the following
wizard is shown: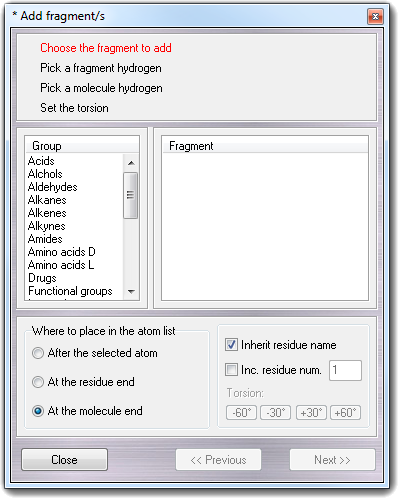
![]() Add
Add
![]() Hydrogens menu item:
Hydrogens menu item: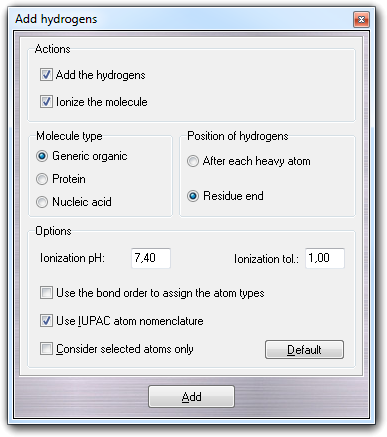
![]() Add
Add
![]() Cluster
in
the main menu, you can solvate a molecule with any type of solvent. The list of solvent is
dynamically created, reading the ...\VEGA ZZ\Data\Clusters directory and can be refreshed closing
and reopening the dialog window. Click here for more
information about the cluster file format.
Cluster
in
the main menu, you can solvate a molecule with any type of solvent. The list of solvent is
dynamically created, reading the ...\VEGA ZZ\Data\Clusters directory and can be refreshed closing
and reopening the dialog window. Click here for more
information about the cluster file format.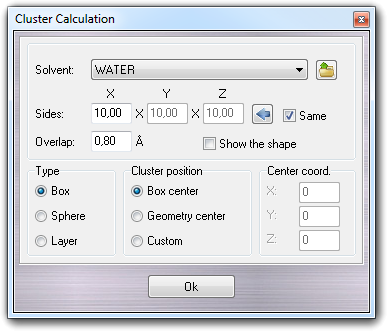
 button. The shape type (Box, Sphere and Layer)
influences the meaning of Sides fields. If Type is Box,
you can put the solvent box dimensions in ┼ngstr÷m. If Same is checked,
the box will be cubic. If Type is Sphere, you can put the Radius
of the cluster sphere in ┼ngstr÷m. If Type is Layer, you
can specify the thickness of the solvent in ┼ngstr÷m. Overlap
parameter indicates the maximum overlapping of the solvent-solute Van der Waals
spheres. In Cluster position you can choose where the cluster is placed
at the center of the box including the solute (Box center) or at the
barycentre of the solute (Geometry center ). When you select Custom,
you can specify the coordinates of the cluster centre. This feature is
useful when you have to solvate with an asymmetric cluster (e.g. phospholipidic bilayer)
and you want to place the solute in the middle. Checking Show the shape,
a preview of the solvent shape is shown making easier the cluster setup.
Clicking
button. The shape type (Box, Sphere and Layer)
influences the meaning of Sides fields. If Type is Box,
you can put the solvent box dimensions in ┼ngstr÷m. If Same is checked,
the box will be cubic. If Type is Sphere, you can put the Radius
of the cluster sphere in ┼ngstr÷m. If Type is Layer, you
can specify the thickness of the solvent in ┼ngstr÷m. Overlap
parameter indicates the maximum overlapping of the solvent-solute Van der Waals
spheres. In Cluster position you can choose where the cluster is placed
at the center of the box including the solute (Box center) or at the
barycentre of the solute (Geometry center ). When you select Custom,
you can specify the coordinates of the cluster centre. This feature is
useful when you have to solvate with an asymmetric cluster (e.g. phospholipidic bilayer)
and you want to place the solute in the middle. Checking Show the shape,
a preview of the solvent shape is shown making easier the cluster setup.
Clicking
![]() button, the cluster dimensions are automatically calculated on the basis of the
solute size. This function isn't available if the shape type is set to Layer.
button, the cluster dimensions are automatically calculated on the basis of the
solute size. This function isn't available if the shape type is set to Layer.
![]() Add
Add
![]() Ions in the
main menu, you can add one or more counter ions to the active molecule:
Ions in the
main menu, you can add one or more counter ions to the active molecule: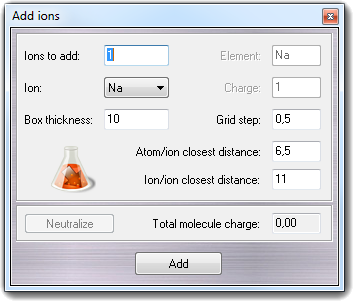
![]() Select
Select
![]() Custom)
and Edit
Custom)
and Edit
![]() Remove
Remove
![]() Invisible atoms menu item.
Invisible atoms menu item.![]() Remove
Remove
![]() Molecule menu item:
Molecule menu item: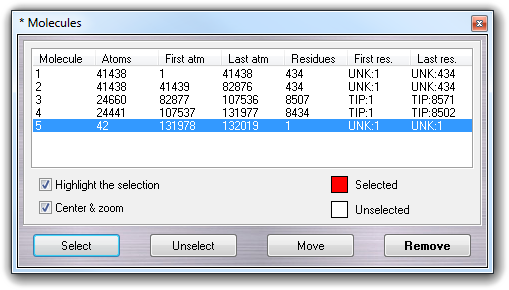
![]() Remove
Remove
![]() Segment menu item:
Segment menu item: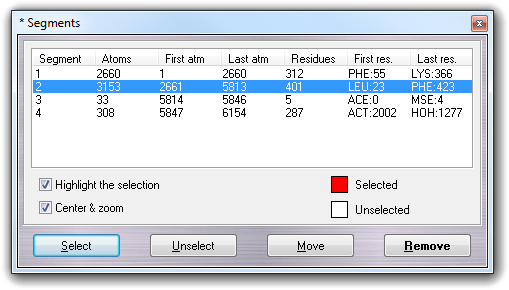
![]() Remove
Remove
![]() Residue main menu item:
Residue main menu item: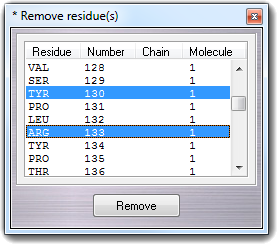
![]() Build
Build
![]() DNA/RNA in the main menu.
DNA/RNA in the main menu.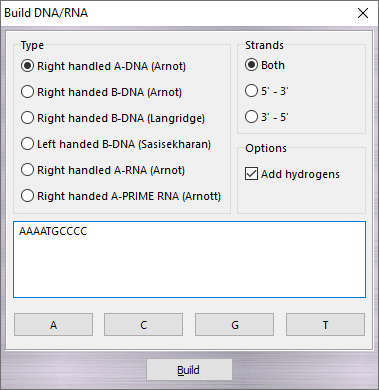
![]() 3' or 3'
3' or 3'
![]() 5') or double (Both) strand, while checking Add hydrogens, the
hydrogens are automatically added to the model.
5') or double (Both) strand, while checking Add hydrogens, the
hydrogens are automatically added to the model.![]() Build
Build
![]() Peptide in the main menu. The build procedure consists in two phases: 1)
construction of the linear peptide; 2) change of the secondary structure.
Peptide in the main menu. The build procedure consists in two phases: 1)
construction of the linear peptide; 2) change of the secondary structure.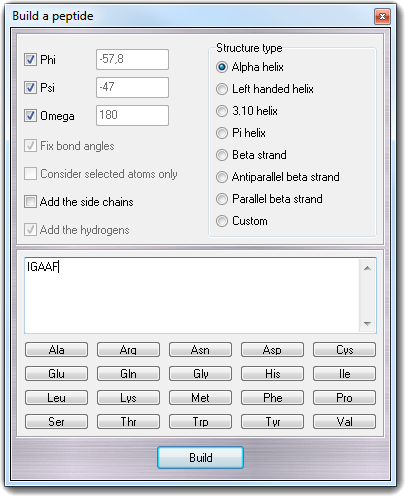

![]() Build
Build
![]() Cluster menu item, it 's possible
to build a solvent cluster useful for the solute solvation (for more information,
click here). In order to create a new solvent
cluster usable by VEGA ZZ, you should follow these steps:
Cluster menu item, it 's possible
to build a solvent cluster useful for the solute solvation (for more information,
click here). In order to create a new solvent
cluster usable by VEGA ZZ, you should follow these steps: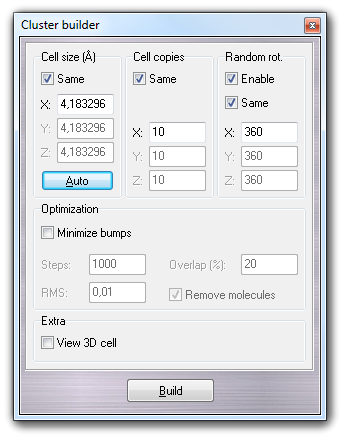
![]() Molecule item in the popup menu.
Molecule item in the popup menu.
![]() Build
Build
![]() SMILES in the main menu.
The 1D structure of the SMILES string is converted to 3D clicking the Build
button. You can select Current workspace or New workspace
as Destination of the 3D structure.
SMILES in the main menu.
The 1D structure of the SMILES string is converted to 3D clicking the Build
button. You can select Current workspace or New workspace
as Destination of the 3D structure.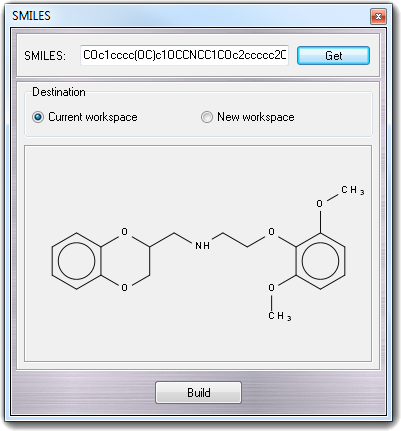
![]() Copy special in the main
menu and by choosing SMILES, but the resulting string is copied to the
clipboard instead of the SMILES window.
Copy special in the main
menu and by choosing SMILES, but the resulting string is copied to the
clipboard instead of the SMILES window.![]() Build
Build
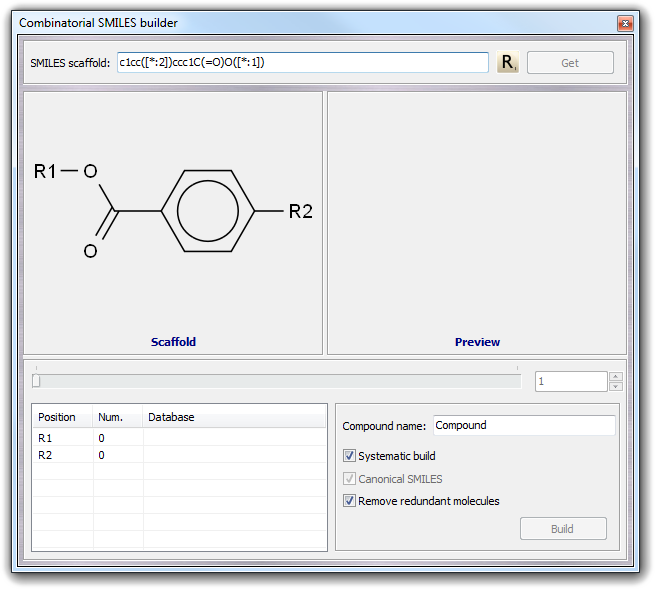
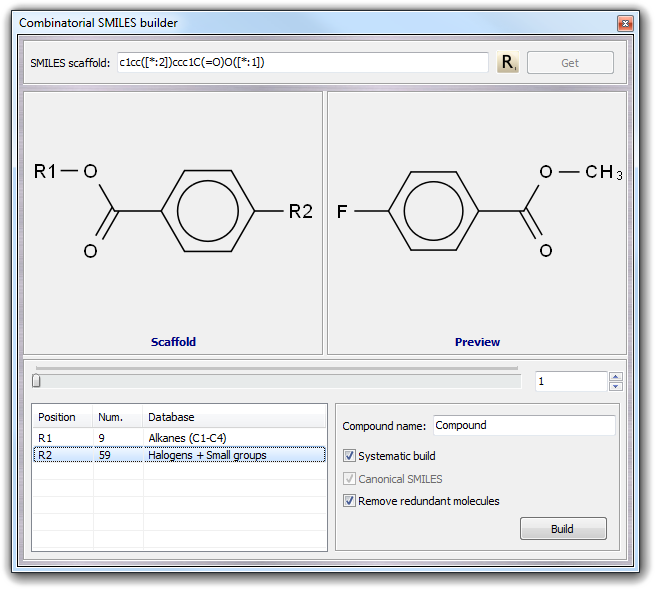
![]() Combi SMILES in the main menu. As in the standard SMILES builder, you can
put directly the scaffold structure in the SMILES scaffold field or you
can obtain it from the current workspace by clicking Get button. For
example, imagine to build a set of derivatives of the benzoic acid:
Combi SMILES in the main menu. As in the standard SMILES builder, you can
put directly the scaffold structure in the SMILES scaffold field or you
can obtain it from the current workspace by clicking Get button. For
example, imagine to build a set of derivatives of the benzoic acid: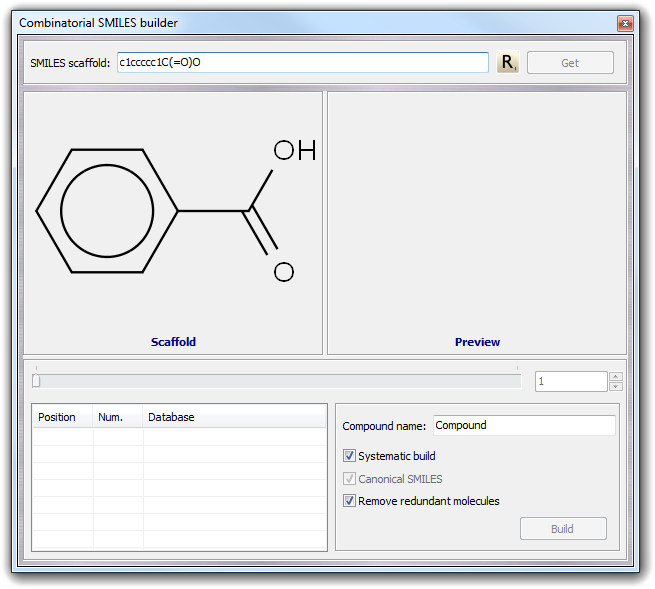
![]() Build
Build
![]() IUPAC in the main menu.
The structure is built and converted to 3D by clicking the Build
button. You can select Current workspace or New workspace
as Destination of the 3D structure.
IUPAC in the main menu.
The structure is built and converted to 3D by clicking the Build
button. You can select Current workspace or New workspace
as Destination of the 3D structure.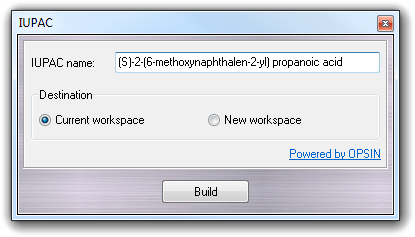
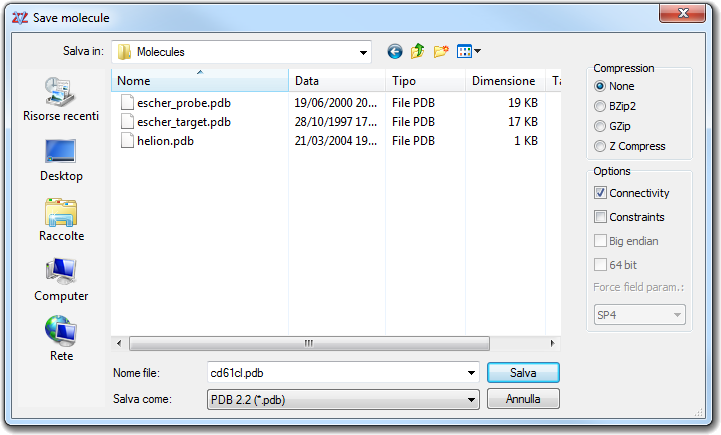
![]() Save main menu item or click the disk picture of the Tool
bar 1. You can choose the File name
and the file format (Save as), but you can also select the Compression
mode
(None, BZip2, GZip and Z Compress) and the possibility to save the Connectivity
and the atom Constraints for the molecular dynamics. Two file formats
only are able to store constraint data and they are PDB (in the
B column for NAMD) and IFF/RIFF. The Big endian and 64 bit
checkboxes are specific for IFF/RIFF format. The former allows to write
in big endian format (it's the real IFF written by old VEGA releases) and
the latter switches the saver in 64 bit mode in order to write files larger
than 4 Gb.
Save main menu item or click the disk picture of the Tool
bar 1. You can choose the File name
and the file format (Save as), but you can also select the Compression
mode
(None, BZip2, GZip and Z Compress) and the possibility to save the Connectivity
and the atom Constraints for the molecular dynamics. Two file formats
only are able to store constraint data and they are PDB (in the
B column for NAMD) and IFF/RIFF. The Big endian and 64 bit
checkboxes are specific for IFF/RIFF format. The former allows to write
in big endian format (it's the real IFF written by old VEGA releases) and
the latter switches the saver in 64 bit mode in order to write files larger
than 4 Gb.![]() Merge menu
item, you can select the file to merge (source) with the molecule already
in memory (target). In the dialog box, you can choose the fields
included in the source molecule that will be read to be merged with the target:
Merge menu
item, you can select the file to merge (source) with the molecule already
in memory (target). In the dialog box, you can choose the fields
included in the source molecule that will be read to be merged with the target: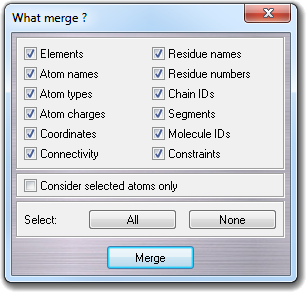
![]() Save Trajectory in the main menu:
Save Trajectory in the main menu: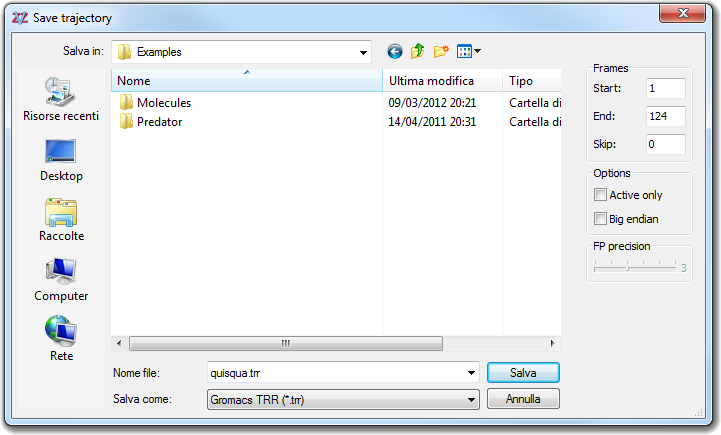
![]() Remove
Remove
![]() Invisible atoms) and save the
reference molecule file with the number of atoms compatible with the new
trajectory (File
Invisible atoms) and save the
reference molecule file with the number of atoms compatible with the new
trajectory (File
![]() Save as ...).
Save as ...).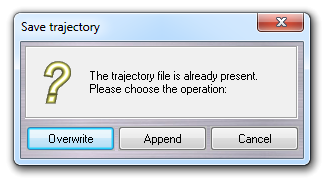
![]() Open menu item.
Open menu item.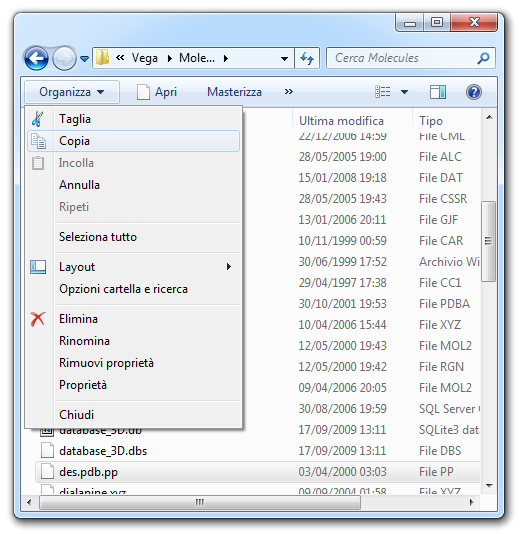
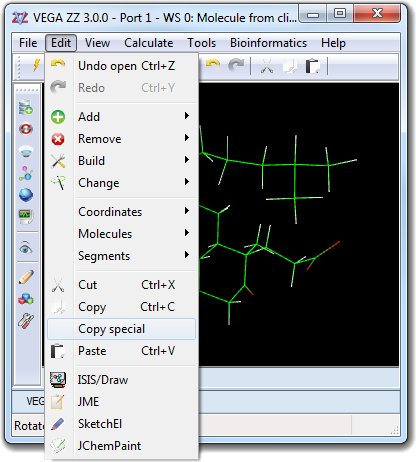
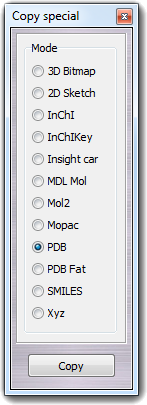
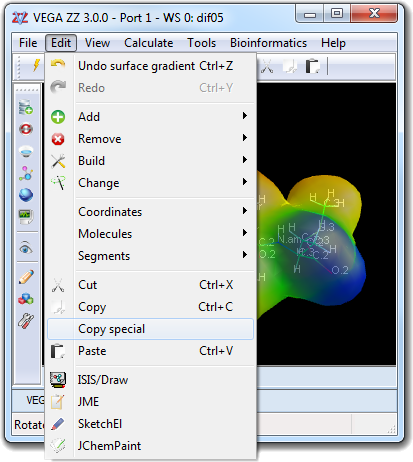
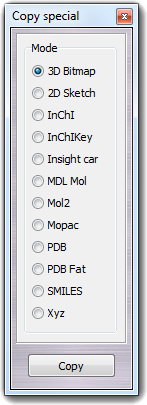
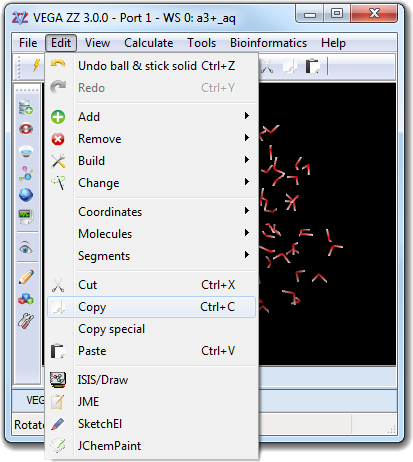
![]() Select in order to change the properties of the visible atoms
only (e.g. color,
measure, etc). If the predefined selections are insufficient, it's possible to create a
custom selection, that can be made by atom specification, by range, by
proximity and slice. This dialog box can be opened selecting the View
Select in order to change the properties of the visible atoms
only (e.g. color,
measure, etc). If the predefined selections are insufficient, it's possible to create a
custom selection, that can be made by atom specification, by range, by
proximity and slice. This dialog box can be opened selecting the View
![]() Select
Select ![]() Custom item in the main menu.
Custom item in the main menu.![]() and
and
![]() adds/removes the atoms using the selected criteria. The Hydrognens, Backbone
and Water buttons are for fast selections: the first one selects all
hydrogens, the second one selects the backbone atoms and the third one selects the water
molecules. Checking Invert, the meaning of the above buttons is inverted
(e.g. clicking Hydrogens, all hydrogen atoms are unselected). The Display box contains controls useful
to change the visualization in selective way: you can change the visualization
mode (Don't change, Wireframe, CPK dotted, CPK wireframe,
CPK solid, Ball &
stick wireframe, Ball & stick solid, Stick wireframe, Stick solid,
Tube and
Trace) and the color of groups of atoms. This last option is enabled only if Change
color is checked. For more information, see the Display
modes section.
adds/removes the atoms using the selected criteria. The Hydrognens, Backbone
and Water buttons are for fast selections: the first one selects all
hydrogens, the second one selects the backbone atoms and the third one selects the water
molecules. Checking Invert, the meaning of the above buttons is inverted
(e.g. clicking Hydrogens, all hydrogen atoms are unselected). The Display box contains controls useful
to change the visualization in selective way: you can change the visualization
mode (Don't change, Wireframe, CPK dotted, CPK wireframe,
CPK solid, Ball &
stick wireframe, Ball & stick solid, Stick wireframe, Stick solid,
Tube and
Trace) and the color of groups of atoms. This last option is enabled only if Change
color is checked. For more information, see the Display
modes section.![]() Display item of the main menu
or Display
item of the context menu or the atom
selection dialog box. You can switch between wireframe,
CPK dotted, CPK wireframe, CPK solid, ball & stick wireframe, ball &
stick solid, stick wireframe, stick solid, trace and tube visualizations.
Display item of the main menu
or Display
item of the context menu or the atom
selection dialog box. You can switch between wireframe,
CPK dotted, CPK wireframe, CPK solid, ball & stick wireframe, ball &
stick solid, stick wireframe, stick solid, trace and tube visualizations.




![]() Display
Display
![]() Settings item of the main menu or the
Display
Settings item of the main menu or the
Display
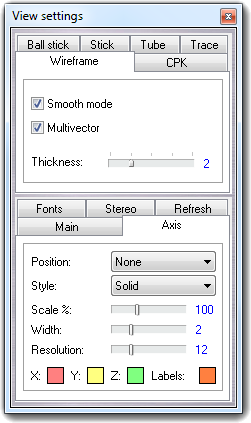
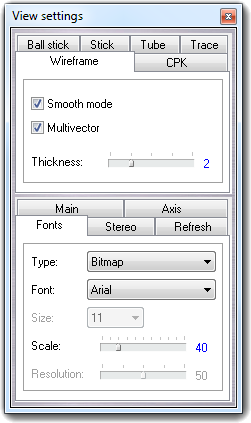
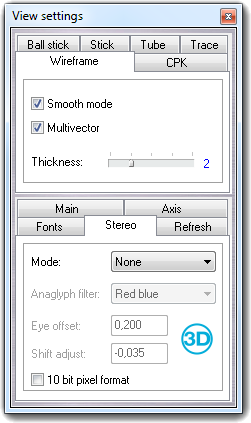
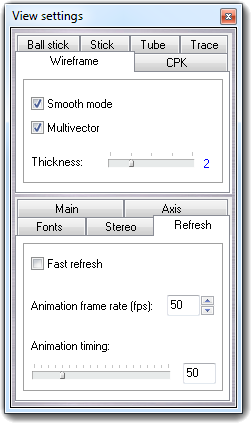
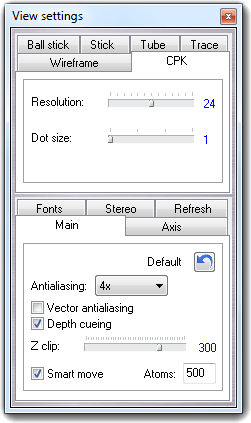
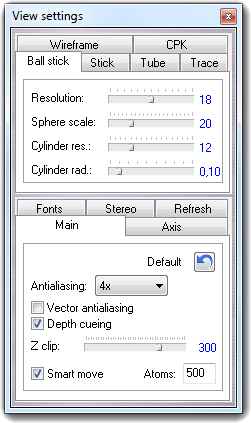
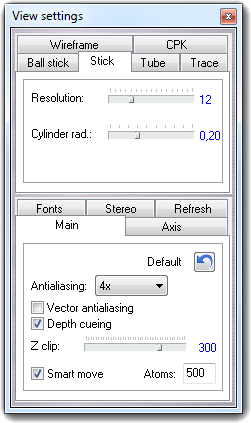
![]() Settings item of the context menu. Press the Done
button to close the dialog.
Settings item of the context menu. Press the Done
button to close the dialog.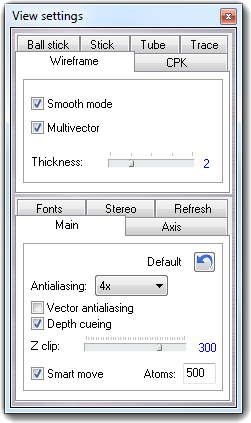





![]() Color menu (click here
for more information). The coloring methods are:
Color menu (click here
for more information). The coloring methods are:![]() Color
Color
![]() Selection
menu item, it's possible to choose the color of the visible atoms from the color table, as
shown below:
Selection
menu item, it's possible to choose the color of the visible atoms from the color table, as
shown below:
![]() Color
Color
![]() Settings, it's possible to change some
interesting color parameters for a nice view.
Settings, it's possible to change some
interesting color parameters for a nice view. In
this tab, you can change the color of the H-bond acceptors, H-bond donors
and non H-bond involved atoms. Moreover, you can change the color of
flexible and rigid bonds.
In
this tab, you can change the color of the H-bond acceptors, H-bond donors
and non H-bond involved atoms. Moreover, you can change the color of
flexible and rigid bonds.![]() Light in the
main menu and allows the light source management.
Light in the
main menu and allows the light source management.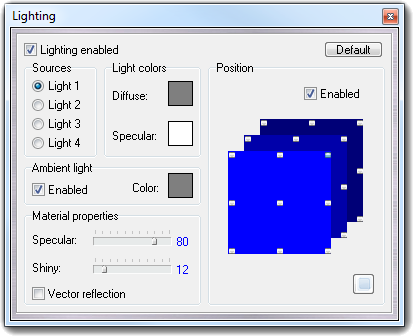
![]() button. For each light source, in the Light colors box, you can change the diffusion and the specularity colors.
You can also switch on/off the ambient light and change its color (see Ambient
light box).
button. For each light source, in the Light colors box, you can change the diffusion and the specularity colors.
You can also switch on/off the ambient light and change its color (see Ambient
light box).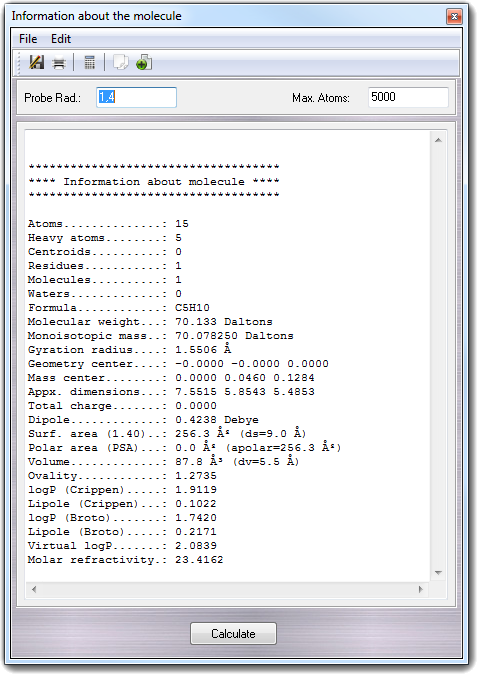
![]() Information
from the main menu, the VEGA ZZ shows a dialog box in which it's possible to set the probe
radius for surface calculations and the maximum number of allowed atoms for time-expensive
calculations (e.g. volume, surfaces, Virtual logP, etc). To perform all
calculations, you must click Calculate button or select Calculate
item from File menu.
Information
from the main menu, the VEGA ZZ shows a dialog box in which it's possible to set the probe
radius for surface calculations and the maximum number of allowed atoms for time-expensive
calculations (e.g. volume, surfaces, Virtual logP, etc). To perform all
calculations, you must click Calculate button or select Calculate
item from File menu. ![]() Hydrogen bonds in
the context menu (press right button on the 3D visualization area):
Hydrogen bonds in
the context menu (press right button on the 3D visualization area): 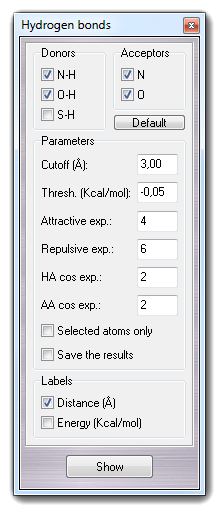
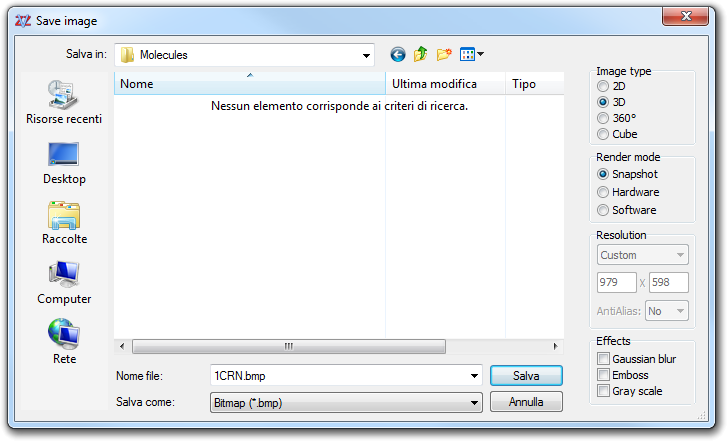
![]() Save image in main menu or
click the camera picture of the Tool bar 1:
Save image in main menu or
click the camera picture of the Tool bar 1: