Viale Abruzzi, 42 - I-20131 Milano (Italy)
A. Pedretti, G. Vistoli, A.M. Villa, L. Villa
 |
Istituto di Chimica Farmaceutica e
Tossicologica - Milan University Viale Abruzzi, 42 - I-20131 Milano (Italy) |
INTRODUCTION
Farnesyl protein
transferase (FTase) catalyzes the transfer of a farnesyl
group from farnesyl diphosphate (FPP) to a specific
cysteine residue of a substrate protein through covalent
attachment (3,21).
This enzyme, like as geranylgeranyl-transferase, recognizes a
common CA1A2X
amino acid sequence (6) located at the
C-terminus of substrate proteins. In the CA1A2X
motif, C is the cysteine residue to which the prenyl
group is attached, A1 and A2
are aliphatic amino acids, and X is the carboxyl
terminus that specifies which prenyl group is attached. If X
is Ala, Cys, Gln, Met, or Ser, the protein is a substrate for
FTase and is farnesylated. If X is Leu or Phe, the
protein is geranylgeranylated. This post-translational
modification is believed to be involved in membrane association
due to the enhanced hydrophobicity of the protein upon
farnesylation. This modification process has been identified in
numerous proteins located in eukaryotic organisms, including Ras
proteins. Ras proteins play a crucial role in the signal
transduction pathway that leads to cell division. It has been
shown that farnesylation of Ras is necessary for proper
functioning in cell signaling.
Recently, there has been widespread interest in studying protein
prenylation since Ras oncoproteins are farnesylated and mutant
forms of Ras have been detected in 30% of human cancers. Since
the farnesylation of oncogenic Ras proteins is required for
cellular transformation, preventing the farnesylation process may
be a possible approach for cancer chemotherapy. This prevention
may be achieved through developing specific inhibitors of FTase,
the enzyme that catalyzes the farnesylation of Ras; the design of
such FTase inhibitors is currently a major area of research.
Knowledge about the active site environment of FTase is important
for designing new, potent inhibitors of the enzyme. Recently the crystal structure of
rat FTase was resolved at 2.25 Å
resolution (15). This protein is
an heterodimer consisting of 48 kD (alpha) and 46 kD (beta)
subunits (6) and the secondary structure of both the and
subunit appear largely composed of alpha helices (4, 5). A single zinc ion,
involved in catalysis, is located at junction between the
hydrophilic surface of beta subunit and a hydrophobic deep cleft
of alpha subunit. The zinc is coordinated by the beta subunit
residues Asp-297, Cys-299, His-362 and a water molecule (6).
Cross-linking studies indicate that the binding sites for both
protein and FPP reside on the subunit (8). The location for the two substrates can be
inferred from the presence of two clefts that differ in their
surface properties. One cleft is hydrophilic, being lined with
charged residues and interacts with the CAAX peptide. The other
cleft, orthogonal to this peptide bindig site, is hydrophobic,
being lined with aromatic residues and is considered the site of
FPP binding (7, 8).
RAS PROTEIN POSTTRANSLATIONAL MODIFICATIONS
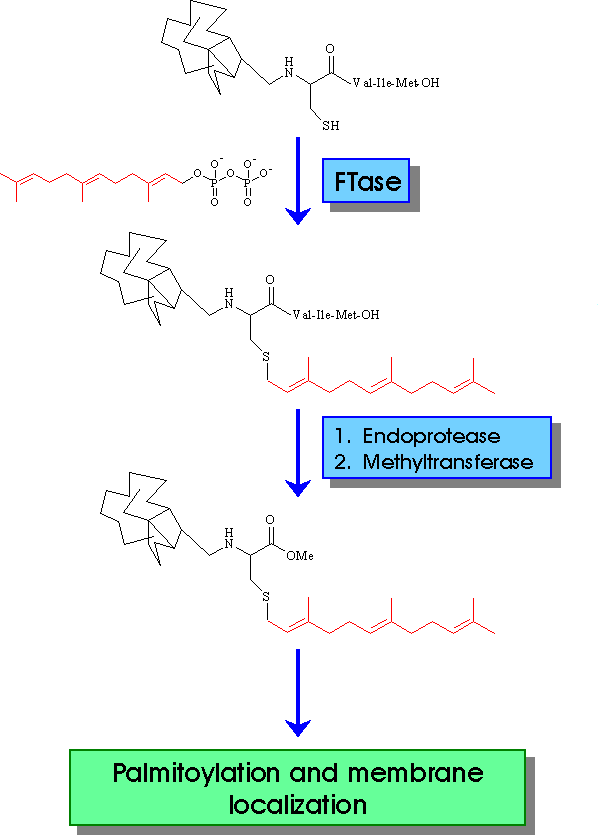
The first step of RAS protein posttranslational modification is the covalent linkage between FPP, derived by classical isoprenoid biosynthesis pathway, and cysteine residue of CAAX (1, 2). This step is followed by cleavage of the last three aminoacids. The identification of the protein responsible for the proteolytic cleavage offers another target for blocking RAS activation. The final posttranslational modification, prior to membrane anchorage, is the methylation of the carboxyl group of prenylated cysteine. S-adenosyl-L-methionine (AdoMet) is the methyl donor. Inhibitors against the methyltransferase has been reported. The next modification is the palmitoylation of cysteine residue located upstream of farnesylated cysteine. This modification increases the binding affinity to the cell membrane, although not be essential.